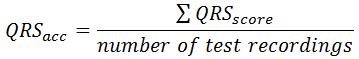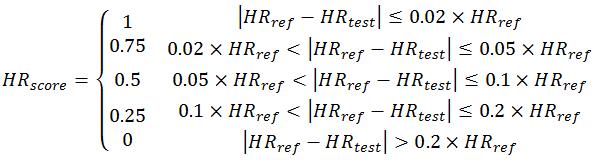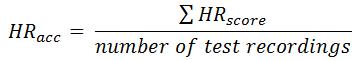If you use the Challenge data for paper publication, please cite this paper for Challenge data description:
H. X. Gao, C. Y. Liu
∗, X. Y. Wang, L. N. Zhao, Q. Shen, E. Y. K. Ng, and J. Q. Li. An Open-Access ECG Database for Algorithm Evaluation of QRS Detection and
Heart Rate Estimation. Journal of Medical Imaging and Health Informatics, 2019, 9(9): 1853–1858.
 The final code is released as
open-source data
The final code is released as
open-source data.
In order to protect the time-effectiveness of each team's algorithm, the organizing committee will postpone opening the trained model for a few months. Thank you for your kind understanding. [December 4, 2019]
The final challenge scores of CPSC 2019 have been indicated as below:
| Code No. | Type | Institution/Affiliation | Team Members | Final score |
| QRSacc | HRacc |
| CPSC0416 | Python | University of Shanghai for Science and Technology | Wenjie Cai, Danqin Hu, Liangyun Jiang | 0.9214 | 0.9489 |
| CPSC0433 | Python | Soochow University | Lishen Qiu, Wenqiang Cai, Miao Zhang, Wanyue Li, Jie Yu, Lesong Zheng, Mixue Deng, Lirong Wang | 0.9155 | 0.9429 |
| CPSC0413 | Python | Wuhan Zoncare Bio-medical Electronics Co., Ltd | Tao Zhu, Yao Wang, Quan Fang | 0.9149* | 0.9431* |
| CPSC0436 | Python | University of Electronic Science and Technology of China | Jin Qi, Sunfeng Luo | 0.9079 | 0.9377 |
| CPSC0430 | Python | Chengdu Spaceon Electronics CO.,LTD. | Chunli Wang, Shan Yang, Xun Tang, Bin Li | 0.9004 | 0.9421* |
| CPSC0437 | Python | Harbin Institute of Technology | Runnan He, Yang Liu, Na Zhao | 0.8921 | 0.928 |
| CPSC0423 | Python | Wuhan Zoncare Bio-medical Electronics Co., Ltd | Yi Li, Jiabin Zhu, Wei Luo | 0.8808 | 0.9239 |
| CPSC0427 | Python | Healthcare Technology Innovation Centre, IIT-Madras, India | Sricharan Vijayarangan | 0.8745 | 0.9261 |
| CPSC0343 | Python | East China Jiaotong University | Lingfeng Liu, Baodan Bai, Xinrong Chen | 0.8664 | 0.9173 |
| CPSC0438 | Python | Zhejiang university | Rongqin Chen, Han Shao, Zuo Wang | 0.8505 | 0.8994 |
| CPSC0401 | Python | Southeast University | Borui Hou, Xin Zhang | 0.8309 | 0.8942 |
| CPSC0434 | Python | Chongqing University of Posts and Telecommunications | Sijia He, Meiqiu Jiang, Xuesha Mo | 0.8240 | 0.8905 |
| CPSC0328 | Python | Shenzhen Digital Life Institute | Qixun Qu | 0.8158 | 0.8756 |
| CPSC0370 | Python | Chengdu Spaceon Electronics CO., LTD. | Peter | 0.8093 | 0.8976 |
| CPSC0332 | Python | Chengdu Lanjing Data&Information Technology Co.,LTD | Yuan Zhuo, Wei Han, Jifan Zhao | 0.8083 | 0.8827 |
| CPSC0348 | Python | Guangzhou Shiyuan Electronics Co., Ltd | Wei Zhao, Jing Hu, Dongya Jia, Hongmei Wang | 0.8002 | 0.8793 |
| CPSC0422 | Python | Qilu University of Technology | Zhongyi Jin | 0.7477 | 0.8702 |
| CPSC0316 | Python | China Medical University | Shuo Zhang, Ruiqing Zhang | 0.6958 | 0.8379 |
| CPSC0383 | Python | Anhui Normal University | Songsong Yan, Jiajia Wang, Yongtao Wang, Lei Qiu | 0.6628 | 0.7997 |
| CPSC0304 | Python | Southeast University | Yang Qin, Lingfei Mo | 0.6511 | 0.7432 |
| CPSC0296 | Python | Guangdong University of Technology | Jie Luo, Jun Lv, Xiangyu Li, Yachun Zheng, Shengli Xie | 0.6064 | 0.6541 |
| CPSC0432 | Matlab | Ludong Univeristy | Wei Li, Laiguo Li, Chunhua Wang, Qian Li, Saiai Bu | 0.5732 | 0.7375 |
| CPSC0428 | Python | Taiwan AI Academy; Academia Sinica | Tsai-Min Chen, Chih-Han Huang, Jhih-Yu Chen, Edward S. C. Shih, Kuo-Hsuan Hung, Heng-Cheng Kuo and Yu Tsao | 0.4240 | 0.6965 |
| CPSC0309 | Matlab | Beijing University of Posts and Telecommunications | Xiuzhu Yang, Mengyao Yang, Yibo Yu, Mu Shen, Hongyu Qian, Xinyue Zhang, Yi Ding, Lin Zhang | 0.4073 | 0.5616 |
| CPSC0311 | Matlab | Beijing Institute of Technology | Chuanbin Ge, Yi Xin, Bingshuai Liu, Qiang Zhang, Shuangfei Li | 0.3743 | 0.5673 |
| CPSC0259 | Matlab | Tsinghua University | Jianping Lin, Fan Yang, Jiawei Li | 0.3442 | 0.5240 |
| CPSC0429 | Python | Ludong University | Xiaoyuan Bian, Chunjian Yang, Fei Niu, Jing Li, Xiaochen Wei | 0.2798 | 0.5602 |
| CPSC0289 | Python | Zhengzhou University | Chuang Han, Hongyi Li, Longlong Qian | 0.1557 | 0.6295 |
| CPSC0307 | Matlab | ChongQing University | Tingting Yu, Wei Qi, Dong Zheng | No score | No score |
*Not engaged in Awarding in case of No Attendance at the conference.
*Please be noted the submission deadline of the final entry
During Oct. 1, 00:00 a.m. - Oct. 7, 12:00 p.m., each team has the opportunity to submit a best entry once, which will be taken as the final entry to be evaluated.
In case of no new entry submitted during the period mentioned above, the entry with the highest score of the team before will be acted as the final version automatically, and run on the whole test database to generate the final challenge results and ranking.
Notes:
We have noticed that all entries estimate HR from QRS locations, however, HR estimation is an independently awarded event which could be improved by sophisiticated algorithms without QRS information.
[2019-9-3] Please download the new
Sample MATLAB entry and
Sample Python entry. The new evaluation method refines the search edge for answers within [0.5, 9.5]s, eliminating the misjudgment of scores while the predictive R peak location is in [0.5, (0.5+0.075)]s or [(9.5-0.075), 9.5]s.
[2019-6-21] To solve the confusion caused by the old version of evaluation algorithm while dealing with the boundary problem, we fixed and resubmit all scoring code. Meanwhile, some labeling error in training set was fixed and resubmit as
Training set!
[2019-5-31] Participants are required to indicate the version of Python or Matlab and the required packages in the Note box during submission.
[2019-5-14] The Sample Python entry is updated for your reference.
[2019-3-26] The last version algorithm of evaluation missed some vital QRS information around the start (0.5s) and end (9.5s) of ECG series, which may result in a lower score. Now we refine it, please download the new Sample MATLAB entry for your scoring.
[2019-3-21] The last 50 data annotations mismatched with recordings because of the overlap in dataset, we have corrected the errors and the training data files in Quick Start have been updated on March 21. Please download the new training set for your code preparation.
The score lists for the first stage has come out as shown below. For those entries which have not been indicated, please check your email to see the error report. Participants are required to indicate the version of Python or Matlab and the required packages in the Note box during submission. Should there be any questions, please feel free to contact
chinachallenge_cpsc@icbeb.org.
Frequently Asked Questions (FAQs)
Q1: How can we register team information to join the challenge?
R1: We suggest that the team leader, or one of the team representatives registers an account on behalf of the team in the system.
Q2: The sample entry, ‘CPSC2019_challenge.m’, is unable to be downloaded.
R2: Right click, save link as, and then download is finished.
Q3: How should I submit the challenge entries with the provided sample entry ‘CPSC2019_challenge’?
R3: You can add the ECG signal processing codes in the ‘CPSC2019_challenge.m', and submit the updated version of 'CPSC2019_challenge.m'. At this moment, please focus on the methods you need to develop, and submit the ‘CPSC2019_challenge.m’ with developed codes. Test data and training data are exactly in the same format.
Q4: How should I name the main function?
R4: Please name it CPSC2019_challenge.
Q5: If we use the python environment, can the test system support commonly used packages, such as keras, wfdb, numpy?
R5: Yes, It can.
Introduction
The China Physiological Signal Challenge 2019 (CPSC 2019) aims to encourage the development of algorithms for challenging QRS detection and heart rate (HR) estimation from short-term single-lead ECG recordings usually with low signal quality and/or abnormal rhythm waveforms.
ECG signal provides an important role in non-invasively monitoring and clinical diagnosis for cardiovascular disease (CVD). Detection of QRS complex is an essential step for ECG signal processing, and can benefit the following HR calculation and abnormal situation analysis. Although detection methods of QRS complex have been severely tracked throughout the last several decades, accurate QRS location and HR estimation are still challenging in noisy signal episode or abnormal rhythm waveforms, especially when the ECG recordings are from the wearable dynamic ECG acquisition. It is true that many of the developed QRS detection algorithms can achieve high accuracy (over 99% in sensitivity and positive predictivity) when tested over the standard ECG databases such as MIT-BIH Arrhythmia Database or AHA Database [1]. However, these algorithms may not be able to perform well when used in the daily life environment that will cause severe noises and significantly reduce the signal quality. A recent study confirmed that none of the common QRS algorithms can obtain 80% detection accuracy when tested in a common dynamic noisy ECG database [2]. Thus, in this challenge, we provide a new ECG database containing noisy ECG episodes and/or signals with different arrhythmia patterns, encouraging participants to develop more efficient and robust algorithms QRS detection and HR estimation. In addition, it is worth to note that, although HR can be calculated from the detection results of QRS complexes, HR can be still estimated without QRS detection step [3,4].
Challenge Data
Training data consists of 2,000 single-lead ECG recordings collected from patients with cardiovascular disease (CVD), each of the recording last for 10 s. Test set contains similar ECG recordings of same lengths, which is unavailable to public and will remain private for the purpose of scoring for the duration of the Challenge and for some period afterwards. ECG recordings were obtained from multiple sources using a variety of instrumentation, although in all cases they are presented as 500 Hz sample rate here. All recordings were provided in MATLAB format (each including two .mat file: one is ECG data and another one is the corresponding QRS annotation file). Pan &Tompkins (P&T) algorithm [5,6] is also provided as benchmark or comparable algorithm.
Although QRS detection and HR estimation are widely studied by lots of researchers for many years, accurate detection is still really challenging in this Challenge due to the QRS amplitude variation, QRS morphological variation, and occurrence of intense variability in the intervals between beats, different arrhythmias, as well as noises. Figure 1 shows the examples of ECG waveforms in training data.
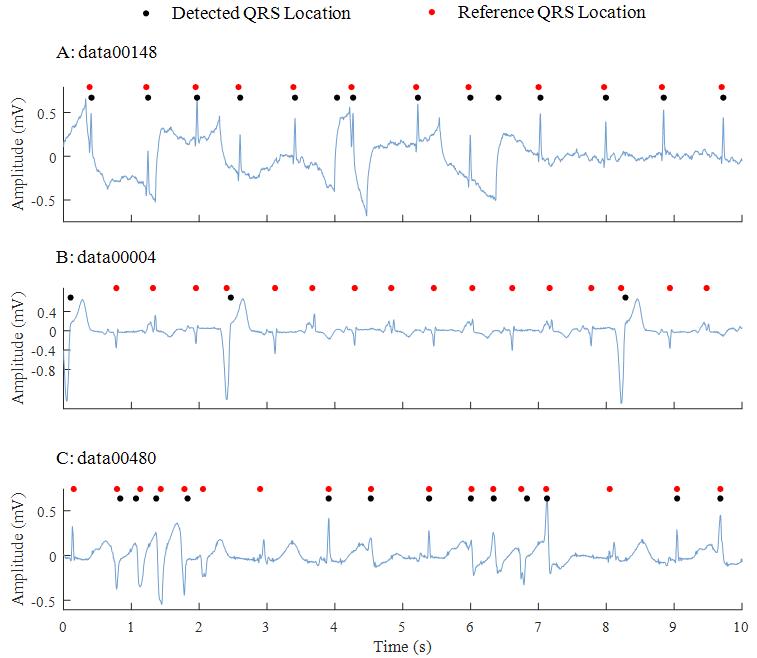 Figure 1.
Figure 1. Examples of the represented ECG waveforms. Red circles denote the reference QRS locations and black ones denote the detected results by the P&T algorithm. The challenges for accurate QRS locations are from: A) intense variability, B) premature ventricular contraction (PVC) and C) ventricular tachycardia.
Challenge Events and Scoring
CPSC 2019 is comprised of two events related to scoring: QRS location and HR estimation. Only test set will be used for event scoring. QRS annotations in the training and test sets are labeled and approved by cardiologists and trained volunteers. These reference annotations have been processed to derive the reference RR interval time series (the intervals between successive QRS annotations). HR is further derived from the RR interval with a 4 s time window ranging from 5.5 s to 9.5 s in each of 10-s ECG segment.
Event 1: QRS detection
In this event, the goal is to produce a set of QRS annotations that can matches the reference QRS annotations. For each reference QRS annotation, a matched QRS annotation should lie in 75 ms duration centered by the reference QRS annotation [9]. Noted that the reference QRS annotations appear in the first and last half seconds are omitted. As shows in Figure 1, detected QRS must be within 75 ms from the reference ones. For each 10-s ECG segment, the scoring rules are:
• complete matching scores one point
• a false positive (FP) detection scores 0.7 points
• a false negative (FN) detection scores 0.3 points, since from a clinical perspective, missed diagnosis is more serious than misdiagnosis, thus we penalize FN detection here
• other situations score 0

The final score for Event 1 (QRS
acc) can be calculated as:
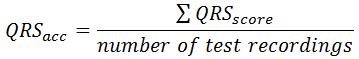 Event 2: HR estimation
Event 2: HR estimation
In this event, the goal is to produce a real-time estimate for HR for a 4-s ECG episode, i.e., ECG signals from 5.5 s to 9.5 s. Figure 2 demonstrates the evaluation method for Event 2, i.e., how HR
score for each 10-s ECG segment can be obtained.
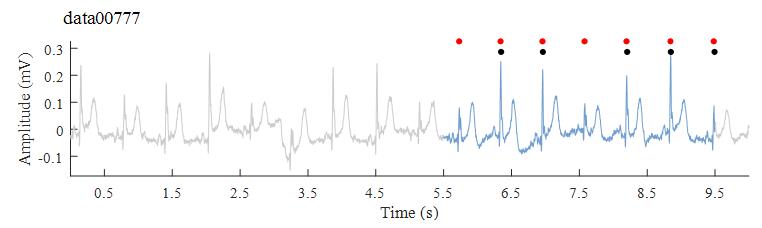 Figure 2.
Figure 2. Demonstration of the evaluation method for Event 2. In this situation, reference HR can be calculated from the reference QRS annotations between 5.5 s and 9.5 s ECG episode (HR
ref = 96), and estimated HR can be calculated from the detected QRS annotations between the same 4-s ECG episode (HR
test = 76).
The detailed scoring rules for HR estimation in each 10-s ECG segment are summarized as:
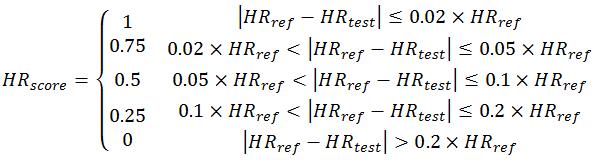
The final score for Event 2 (HR
acc) can be calculated as:
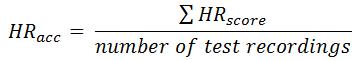
where HR
test and HR
ref represent heart rate calculated by competitor and reference, respectively. HR estimation is usually derived from the QRS detection but we encourage participant to develop robust HR estimation without QRS location.
Quick Start
1. Download the
Training set the
Sample MATLAB entry and
Sample Python entry
(updated on June 21, 2019)
2. Develop your entry by editing the existing files:
• Modify the sample entry source code file CPSC2019_challenge.m with your changes and improvements.
• Modify the AUTHORS.txt file to include the names of all the team members.
• Run your modified source code file on all the records in the training set by executing the script CPSC2019_ccore.m. This will also build a new version of
entry.zip.
• Optional: Include a file named DRYRUN in the top directory of your entry (where the AUTHORS.txt file is located) if you do not wish your entry to be
scored and counted against your limit. This is useful in cases where you wish to make sure that the changes made do not result in any error.
Participating in the challenge
As a starting point, we have provided example entries (
Sample MATLAB entry/
Sample Python entry
) implemented using Matlab or Python, which provides the most widely used QRS peaks annotation algorithm, pan_tompkin.m, as benchmark or comparable algorithm.
To participate in the challenge, you will need to submit a software written by MATLAB or Python that is able to run on the test set and output the final scoring result without user interaction in our test environment. One sample entry CPSC2019_challenge.m is available to help you get started. And a scoring function CPSC2019_score.m is provided for competitors testing algorithms and build a new version of entry.zip. The team must submit the first challenge open-source entry before June 1, 2019. The deadline for submitting the official challenge entries is October 1, 2019.
Awards and Rules
The winner will be selected on the basis of the obtained final QRS
acc and HR
acc on the hidden test data. The first three for each Event challenging will receive certificates and generous bonuses:
• First prize: Certificate plus bonus of RMB 15,000
• Second prize: Certificate plus bonus of RMB 10,000
• Third prize: Certificate plus bonus of RMB 5,000
We welcome all the individual or research group around the world to attend the challenge. To be eligible for the award, please do the following:
(Please be noted that paper submission is not mandatory and will not affect the challenge score.)
(1) Register the team before May 1, 2019.
(2) Submit at least one open-source entry that can be scored before June 1, 2019.
(3) Attend ICBEB 2019 (October 22-25, 2019) in Seoul, Republic of Korea and present your work there.
Please do not submit analysis of this year’s Challenge data to other Conferences or Journals until after ICBEB 2019 has taken place, so the competitors are able to discuss the results in a single forum.
Important Dates
January 25, 2019 -- Challenge open
May 1, 2019 -- Deadline for team registration to join the challenge
June 1, 2019 -- Deadline for submitting the first challenge open-source entry
October 1, 2019 -- Deadline for submitting the challenge entries
October 22-25, 2019 – Announcement of the winners of CPSC 2019 in ICBEB 2019
Any questions or problems about the Challenge, please feel free to contact chinachallenge_cpsc@icbeb.org.
Publication – Call for Chapters
The book with
Springer Nature entitled Feature Engineering and Computational Intelligence in ECG Monitoring calls for chapters.
The purpose of this book is to summarize the feature engineering and computational intelligence solutions for ECG monitoring, with an emphasis on how these methods can be efficiently used on the emerging need and challenge -- dynamic, continuous & long-term individual ECG monitoring and real-time feedback, aiming to provide a “snapshot” of the state of current research at the interface between physiological signal analysis and machine learning. It could help clarify some dilemmas and encourage further investigations in this field, to explore rational applications of feature engineering and computational intelligence in clinical practices for ECG monitoring.
Original, high quality contributions that are not yet published or that are not currently under review by other journals or peer-reviewed conferences are welcomed, with special emphasis on, but not limited to, a number of correlative chapter topics.
Click
Book Proposal for more details.
Challenge Chair:
Chengyu Liu, Southeast University, China
Challenge Committee:
Hongxiang Gao, Southeast University, China
Xingyao Wang, Southeast University, China
Feifei Liu, Southeast University, China
Lina Zhao, Southeast University & Shandong University, China
Xiaoling Wu, Nanjing Medical University, China
International Advisory Chair:
Gari D. Clifford, Emory University & Georgia Institute of Technology, USA
International Advisory Co-chairs:
Aiguo Song, Southeast University, China
Jianqing Li, Nanjing Medical University, China
Zuhong Lu, Southeast University, China
Ye Li, SIAT of Chinese Academy of Sciences, China
Yingjia Yao, Lenovo Group, China
International Advisory Committee:
Eddie Ng Yin Kwee, Nanyang Technological University, Singapore
Amit Shah, Emory University, USA
Yi Peng, Peking Union Medical College, China
Shoushui Wei, Shandong University, China
Zhengtao Cao, Aviation Medicine Institute, China
Hongxing Liu, Nanjing University, China
Fengfeng Zhou, Jilin University, China
Alistair Johnson, MIT, USA
Hosted by:
School of Instrument Science and Engineering, Southeast University, China
The State Key Laboratory of Bioelectronics, Southeast University, China
School of Biomedical Engineering and Information, Nanjing Medical University, China
Supported by:
Health Engineering Branch of Chinese Society of Biomedical Engineering
ICBEB Organizing Committee
Awards sponsored by:
Lenovo Group
Reference
1. G.B. Moody, R.G. Mark, The impact of the MIT-BIH arrhythmia database, IEEE Engineering in Medicine & Biology Magazine the Quarterly Magazine of
the Engineering in Medicine & Biology Society, 20 (2001) 45-50.
2. Liu, F.F.; Wei, S.S.; Li, Y.B.; Jiang, X.E.; Zhang, Z.M.; Liu, C.Y., Performance analysis of ten common qrs detectors on different ecg application cases.
Journal of Healthcare Engineering 2018, 2018, ID 9050812.
3. J.J. Gieraltowski, K. Ciuchcinski, I. Grzegorczyk, K. Kosna, Heart rate variability discovery: Algorithm for detection of heart rate from noisy, multimodal
recordings, Computing in Cardiology, 2014, pp. 253-256.
4. J. Gieraltowski, K. Ciuchcinski, I. Grzegorczyk, K. Kosna, M. Solinski, P.Podziemski, RS slope detection algorithm for extraction of heart rate from noisy,
multimodal recordings, Physiological Measurement, 36 (2015) 1743-1761.
5. P.S. Hamilton, W.J. Tompkins, Quantitative investigation of QRS detection rules using the MIT/BIH arrhythmia database, Biomedical Engineering, IEEE
Transactions on, (1986) 1157-1165.
6. J. Pan, W.J. Tompkins, A real-time QRS detection algorithm, Biomedical Engineering, IEEE Transactions on, (1985) 230-236.
7. ANSI-AAMI (1998). Testing and reporting performance results of cardiac rhythm and st segment measurement algorithms, ANSI-AAMI:EC57.









 The final code is released as
open-source data.
The final code is released as
open-source data.